Cascade screening of relatives for familiar hypercholesterolemia: detection of low density lipoprotein receptor gene mutations using real-time PCR
Early detection of disease-associated mutations in patients with familial hypercholesterolemia (FH) is crucial for early interventions that can reduce the risk of cardiovascular disease. Here, we describe real-time PCR-based approaches for the rapid detection of single nucleotide substitutions or insertions of the low density lipoprotein receptor gene for cascade screening of relatives.
by Sarojini Pandey and Dimitris K. Grammatopoulos
Introduction
Familial hypercholesterolemia (FH) 5 (OMIM#606945) is an autosomal-dominant disorder associated with abnormally high serum concentrations of low density lipoprotein (LDL) cholesterol (LDL-C) [1]. FH is one of the most common inherited disorders, with a worldwide prevalence estimated at 1 in 200–500 [2]. Affected individuals have increased risk of premature coronary heart disease and death [3]; however, most remain undiagnosed, untreated or inadequately treated. It has been proven that early detection of the disease and treatment reduces morbidity and mortality [4]. The majority of FH cases are caused by genetic defects in the LDL receptor (LDLR) as well as apolipoprotein B, or proprotein convertase subtilisin/kexin type 9. More than 80% of FH patients have mutations in the LDLR gene [5]. Over 1400 different mutations are listed in the LDLR gene database of University College London to date.
To address the screening deficit, the National Institute for Health and Clinical Excellence (NICE) in the United Kingdom developed guidelines on FH management strongly recommending identification of causal mutations in suspected cases of FH phenotype and cascade screening of relatives using a combination of genetic testing and LDL-C concentration measurement to identify affected relatives of those index individuals with a clinical diagnosis of FH [6]. This approach of genetic testing of affected individuals and screening of relatives is considered the most cost-effective strategy for detecting cases of FH across the population [7]. However, the most appropriate and cost-effective diagnostic testing protocol for use across the FH clinical diagnostic services remains to be established. Here, we describe an experimental approach suitable for the rapid detection of known single nucleotide substitutions or insertions of the LDLR gene in suspected individuals using real-time based PCR.
Real-time PCR-based method for identifying LDLR gene mutations
Genomic DNA was extracted from saliva or EDTA-containing blood samples using a QIAamp DNA Blood Mini Kit (Qiagen), and DNA concentration was quantified by ND-1000 spectrophotometer (NanoDrop, Thermo Scientific).
Genomic DNA was amplified with specific oligonucleotide primers and fluorescently labelled probes to identify the PCR product (LC FastStart DNA Master Hybridization Probe kit, Roche). The specific genotype was determined by performing a melting-curve analysis based on fluorescence resonance energy transfer (FRET) technique. Each 10-μL reaction contained 1× LightCycler FastStart DNA Master HybProbe, 3 mmol/L MgCl2, 500 nmol/L of forward and reverse primers, and 200 nmol/L of each hybridization probe. The amplification conditions consisted of one denaturation/activation cycle of 10 min at 95 °C and 45 cycles of three-temperature amplification. Each cycle consisted of 95 °C for 10 seconds, 60 °C for 10 seconds, and 72 °C for 15 seconds with a single fluorescence acquisition step at the 60 °C hold. This was followed by a melting-curve analysis of 95 °C for 20 seconds, 40 °C for 20 seconds, and a slow ramp (0.2 °C/second) to 85 °C with continuous fluorescence acquisition [8].
For LDLR 2054C>T genotyping the LightSNP® Kit rs28942084 LDLR [P685L] from TIB MOLBIOL (Berlin, Germany) whereas LDLR c.1474G>A; c.1567G>C; c.487dupC and c.647G>C mutations were identified by custom-made assays as previously described [8].
Results
Repeatability/reproducibility studies using five replicates of the same DNA sample or different batches of DNAs of heterogeneous genotypes were analysed five times and showed no intra-patient or between-batch variation. All LightCycler assays consistently identified the genotype correctly, confirming their analytical reliability and suitability for routine use.
All PCR methods demonstrated excellent robustness and analytical performance characteristics even when processing genomic DNA of less than optimal DNA purity (absorbance ratio 260/280 <1.6) and quantity (2.5–50 ng/μL). The genotype of all patients tested was correctly identified.
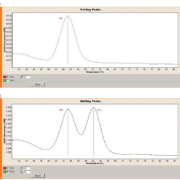
Figure 1 shows examples of wild-type and heterozygous for the LDLR c.1474G>A mutation. Heterozygote patients showed two distinct melting peaks and the G>A nucleotide substitution was detected by a melting temperature (Tm) shift of 7 °C.
In addition to ease of use and cost-effectiveness, a major advantage of this methodology is the rapid turn-around time of 90 min from genomic DNA extraction to PCR genotyping. This identifies potential uses outside large specialist centres in local one-stop clinics.
Discussion
The UK National Institute for Health and Care Excellence (NICE) recommends genetic testing of candidate patients presenting with FH phenotype and, once a disease-causing mutation is identified, screening of relatives; this is considered as the most cost-effective strategy for early detection of unsuspected cases of FH [9], and for distinguishing monogenic FH from sporadic or polygenic hypercholesterolaemia [10]. Detection of unknown mutations in the LDLR gene, where the majority of disease-causing mutations are found, requires complex and specialized molecular methods suitable for comprehensive scanning of the nucleotide sequence [11]. In contrast, once the disease-causing mutation has been identified, screening of relatives for the presence of the mutation does not pose a significant analytical challenge and a number of methodologies are available to the diagnostic services. Selection of these methods ultimately depends on local clinical service configuration, available laboratory expertise and resources and budget constraints. Some of these test requirements can be addressed by real-time PCR methods, which provide a cost-effective (the cost of each PCR method is estimated below £20) and rapid method for screening mutations associated with FH in family studies. Thus, these methods have the potential to deliver the second line of investigations of the FH cascade testing NICE pathway. The fast turn-around time of the method offers a significant advantage allowing the provision of a faster service as well as supporting delivery models such as a one-stop lipid clinic. This would allow the fast-tracking of clinical decision-making and choice of treatment as well as patient convenience, thus offering additional financial savings to the healthcare provider.
References
1. Marks D, Thorogood M, Neil HA, Humphries SE. A review on the diagnosis, natural history, and treatment of familial hypercholesterolaemia. Atherosclerosis 2003; 168: 1–14.
2. Benn M, Watts GF, Tybjaerg-Hansen A, Nordestgaard BG. Familial hypercholesterolemia in the Danish general population: prevalence, coronary artery disease, and cholesterol-lowering medication. J Clin Endocrinol Metab 2012; 97: 3956–3964.
3. Austin MA, Hutter CM, Zimmern RL, Humphries SE. Familial hypercholesterolemia and coronary heart disease: a HuGE association review. Am J Epidemiol 2004; 160: 421–429.
4. Neil A, Cooper J, Betteridge J, Capps N, McDowell I, Durrington P, Seed M, Humphries SE. Reductions in all-cause, cancer, and coronary mortality in statin-treated patients with heterozygous familial hypercholesterolaemia: a prospective registry study. Eur Heart J 2008; 29: 2625–2633.
5. Usifo E, Leigh SE, Whittall RA, Lench N, Taylor A, Yeats C, Orengo CA, Martin AC, Celli J, Humphries SE. Low-density lipoprotein receptor gene familial hypercholesterolemia variant database: update and pathological assessment. Ann Hum Genet 2012; 76: 387–401.
6. Chiou KR, Charng MJ, Chang HM. Array-based resequencing for mutations causing familial hypercholesterolemia. Atherosclerosis 2011; 216: 383–389.
7. Hinchcliffe M, Le H, Fimmel A, Molloy L, Freeman L, Sullivan D, Trent RJ. Diagnostic validation of a familial hypercholesterolaemia cohort provides a model for using targeted next generation DNA sequencing in the clinical setting. Pathology 2014; 46: 60–68.
8. Pandey S, Leider M , Khan M , Grammatopoulos DK. Cascade screening for familiar hypercholesterolaemia: PCR methods with melting-curve genotyping for the targeted molecular detection of apolipoprotein B and low density lipoprotein receptor gene mutations to identify affected relatives. JALM 2016; 02: 109–118.
9. Nherera L, Marks D, Minhas R, Thorogood M, Humphries SE. Probabilistic cost-effectiveness analysis of cascade screening for familial hypercholesterolaemia using alternative diagnostic and identification strategies. Heart 2011; 97: 1175–1181.
10. Talmud PJ, Shah S, Whittall R, Futema M, Howard P, Cooper JA, Harrison SC, Li K, Drenos F, et al. Use of low-density lipoprotein cholesterol gene score to distinguish patients with polygenic and monogenic familial hypercholesterolaemia: a case-control study. Lancet 2013; 381: 1293–1301.
11. Hollants S1, Redeker EJ, Matthijs G. Microfluidic amplification as a tool for massive parallel sequencing of the familial hypercholesterolemia genes. Clin Chem 2012; 58: 717–724.
The authors
Sarojini Pandey1 MSc and Dimitris K. Grammatopoulos*1,2 PhD, FRCPath
1Department of Clinical Biochemistry,
University Hospital Coventry and Warwickshire, Coventry CV2 2DX, UK
2Division of Translational and Systems Medicine, Warwick Medical School,
Coventry CV4 7AL,
UK
*Corresponding author
E-mail: Sarojini.Pandey@uhcw.nhs.uk