Clostridium difficile diagnosis: not always a bed of roses
Clostridium difficile is a leading cause of nosocomial diarrhoea and one of the most common healthcare-associated infections. A dramatic worldwide increase in the incidence of C. difficile infection has occurred over the past two decades with the emergence of hypervirulent strains. Accurate and timely laboratory diagnosis of C. difficile infection is fundamental to ensure patients receive appropriate treatment and proper counter-infection measures are put in place. However, the availability of many commercial tests with different C. difficile targets contributes to uncertainty and controversy around the optimal diagnostic algorithm.
by Dr Guilherme Grossi Lopes Cançado, Prof. Rodrigo Otávio Silveira Silva and Prof. Eduardo Garcia Vilela
Introduction
Clostridium difficile is a major cause of healthcare-associated diarrhoea, and is linked to significant morbidity, economic burden and even mortality. Disagreement between diagnostic tests is an ongoing barrier to clinical decision-making and epidemiological surveillance. With the increasing number and severity of Clostridium difficile infections (CDI) after the emergence of the epidemic BI/NAP1/027 strain in the 2000s, there has been a renewed interest in optimizing the laboratory diagnosis of this infection.
Testing for C. difficile
Cytotoxin neutralization, toxigenic culture and toxin enzyme immunoassay
The two most accurate methods for CDI diagnosis are cytotoxin neutralization (CTN) and toxigenic culture (TC). CTN has long been used as a reference method for C. difficile detection, although different protocols have been proposed. Basically, this technique consists of inoculating a filtrate of a stool suspension into a cell culture (Vero, Hep2, fibroblasts, CHO or HeLa cells) and observing a cytopathic effect (such as cell rounding) as a consequence of disruption of the cell cytoskeleton 24–48 h later. CDI confirmation is obtained by the addition of a specific antiserum directed against C. difficile or against C. sordellii, neutralizing the toxin effects. Studies performed in the last 10 years, however, demonstrated that the sensitivities of CTN protocols range between 60 and 86%, when compared with toxigenic culture. Although culture was found to be the most sensitive method for detecting C. difficile in feces, it is not very specific owing to the possibility of isolating non-toxigenic strains. In order to overcome this issue, it should always be combined with a toxin detection method, such as enzyme immunoassay (EIA), direct cytopathic effect on cell lines or the identification of toxin-related genes by PCR. This notwithstanding, both techniques are time consuming, laborious and require trained personnel, reasons why they are not frequently used in daily practice [1].
In this context, commercial rapid EIA and DNA-based tests are currently the most widely used tools for CDI diagnosis. EIA for toxin detection is fast, cheap, easy to perform and does not require technical training or special equipment: characteristics which have favored its use in low income countries. However, several studies have reported low detection sensitivities of different commercial kits (approximately 50–70%), making toxin EIAs inadequate as standalone tests [2]. Some authors have suggested the analysis of sequential samples from the same patient, but this practice did not significantly increase the positive predictive value of the test and may even amplify the rate of false positives. Furthermore, toxin tests can be falsely negative, which can be due to previous antibiotic treatment, pre-analytic toxin degradation or sampling error (e.g., ileus, fecal dilution). Consequently, toxin EIA may be sufficient and cost-effective as a screening test in clinical settings where there is a low prevalence of CDI, but not for epidemic scenarios.
Detection of glutamate dehydrogenase
Another method that has been used to diagnose CDI is the detection of glutamate dehydrogenase (GDH), a metabolic enzyme constitutionally produced almost exclusively by C. difficile, which is significantly more sensitive than toxin A and B EIAs. We have recently shown that GDH rapid immunoassay presents 100% sensitivity and negative predictive value, compared with the culture-based method, in accordance with order authors [3]. Cheng et al. have advocated the use of GDH in order to improve the diagnostic capacity and control of potential outbreaks of CDI in developing countries, as this test is five to ten times cheaper than molecular assays [4]. In fact, we have recently reported a significant increase in CDI treatment rates in a university hospital of Brazil (from 53.8% to 100%) simply after replacing the toxin EIA by the rapid GDH immunoassay as a screening test [3]. This finding supports the use of GDH in countries with limited economic resources, as it is simple to perform and, unlike PCR, requires no special facilities or personnel qualifications that might restrict its use. Although sensitive, GDH lacks specificity, because it only indicates the presence of the microorganism, instead of toxin production. In this context, a concern with the overdiagnosis and overtreatment of patients with diarrhoea and C. difficile colonization may arise. A negative test can almost rule out CDI and avoid the need for more expensive toxin testing, but a positive GDH assay should preferably be followed by toxin-based diagnostic methods before one can come to a safe conclusion on infection. Asymptomatic carriage of C. difficile occurs in 5–15% of healthy adults, but may be as high as 90% in newborns and healthy infants, and up to 51% in residents in long-term care facilities [5]. In this way, the incorporation of a two-step strategy, including a sensitive organism-based test for screening, such as GDH, followed by a toxin test for confirmation of clinically significant disease, is a reasonable approach. Combined tests including GDH and toxin detection in one easy-to-use cartridge have been recently developed, but several authors have demonstrated limited sensitivity of the toxin component. In our study, the toxin component presented an even lower sensitivity than conventional toxin EIA (50% vs 58%), compared with toxigenic culture [3]. In this way, GDH+/toxin− samples would still have to be submitted to a third test (in a multistep algorithm) to rule out infection, increasing time to diagnosis and health costs. However, some studies have shown that, even without treatment, patients with toxin-negative stool specimens have shorter diarrhoea duration than those with toxin-positive stool specimens. These findings may suggest a limited need for CDI treatment for GDH-positive patients and toxin-negative stool specimens [6].
Molecular testing and rapid diagnosis
In 2009, facing an epidemic of CDI due to hypervirulent C. difficile strains, the US FDA cleared the first commercial molecular test for rapid CDI diagnosis [7]. Shortly afterwards, hospitals in North America and Europe began switching to DNA-based testing strategies as a method of choice for the diagnosis of C. difficile. Nucleic acid amplification testing (NAAT), including rapid testing PCR and loop-mediated isothermal amplification (LAMP), can detect the tcdA/tcdB genes (regulate toxin A/B production) or the tcdC gene (a negative regulator of toxin A and B production) and identify the presence of toxigenic C. difficile in a single step. It has a higher sensitivity (90–95%) and specificity (95–96%) than toxin EIAs and has a rapid turnaround time, but requires specialized equipment and personnel. It is worth noting that the Gene-Xpert® system can also simultaneously indicate the presence of the potentially ‘hypervirulent’ ribotype 027 strain, giving important epidemiological information. Nonetheless, although a PCR assay can identify toxin genes, it cannot detect the presence of toxin. Several studies have shown that toxin−/C. difficile+ patients present shorter duration of symptoms and better outcomes than toxin+/C. difficile+ individuals, demonstrating that this subpopulation may have either mild CDI or colonization, and even not need to be treated. In this way, some authors have questioned the use molecular tests as standalone tests because of the high likelihood of overdiagnosis and overtreatment [8]. However, there may be a role for identifying carriers to prevent transmission and this issue should be better addressed in future studies.
C. difficile testing protocol
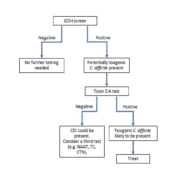
Using reliable and rapid diagnostic tests, such as NAAT, practitioners could offer appropriate treatment earlier, thereby sparing patients a time-consuming evaluation and unnecessary antibiotic therapy and its complications. Despite worldwide advances in analytical technology, transport systems and computerization, many laboratories in developing countries have difficulties in improving turnaround times and diagnostic capability. The optimal diagnostic algorithm for CDI is yet to be adequately defined and may vary according to the underlying clinical and laboratory circumstances. We believe that the decision about which test(s) to use is determined by a combination of what is practical and feasible in a specific setting. In Brazil, for example, where only toxin EIA was available for CDI diagnosis until 2015, the introduction of the GDH test resulted in a dramatic increase in C. difficile treatment rates [3]. All of the reference methods, CTN, toxigenic culture, or PCR, require advanced infrastructure and expensive testing. Smaller, community-based hospitals, where much of C. difficile testing is currently performed, may not have the financial means to establish these methods nor have the staff to perform the time-consuming, highly complex assays. In this way, using GDH–toxin A/B assays may be an adequate option for diagnostic algorithms in developing countries, whereas molecular techniques, toxigenic culture or CTN may be reserved for discordant samples (Figure 1). Future studies should focus on developing simple diagnostic approaches to accurately distinguish active infection from mere colonization.
References
1. Delmée M. Laboratory diagnosis of Clostridium difficile disease. Clin Microbiol Infect 2001; 7(8): 411–416.
2. Silva RO, Vilela EG, Neves MS, Lobato FC. Evaluation of three enzyme immunoassays and a nucleic acid amplification test for the diagnosis of Clostridium difficile-associated diarrhea at a university hospital in Brazil. Rev Soc Bras Med Trop 2014; 47(4): 447–450.
3. Cançado GGL, Silva ROS, Nader AP, Lobato FCF, Vilela EG. Impact of simultaneous glutamate dehydrogenase (GDH) and toxin A/B rapid immunoassay on Clostridium difficile diagnosis and treatment in hospitalized patients with antibiotic-associated diarrhea in a university hospital of Brazil. J Gastroenterol Hepatol 2017; doi: 10.1111/jgh.13901 [Epub ahead of print].
4. Cheng JW, Xiao M, Kudinha T, Xu ZP, Sun LY, Hou X, Zhang L, Fan X, Kong F, Xu YC. The role of glutamate gehydrogenase (GDH) testing assay in the diagnosis of Clostridium difficile infections: a high sensitive screening test and an essential step in the proposed laboratory diagnosis workflow for developing countries like China. PLoS One 2015; 10(12): e0144604.
5. Furuya-Kanamori L, Marquess J, Yakob L, Riley TV, Paterson DL, Foster NF, Huber CA, Clements AC. Asymptomatic Clostridium difficile colonization: epidemiology and clinical implications. BMC Infect Dis 2015; 15: 516.
6. Yuhashi K, Yagihara Y, Misawa Y, Sato T, Saito R, Okugawa S, Moriya K. Diagnosing Clostridium difficile-associated diarrhea using enzyme immunoassay: the clinical significance of toxin negativity in glutamate dehydrogenase-positive patients. Infect Drug Resist 2016; 9: 93–99.
7. Polage CR, Turkiewicz JV, Cohen SH. The never-ending struggle with laboratory testing for Clostridium difficile infection. J Comp Eff Res 2016; 5(2): 113–116.
8. Polage CR, Gyorke CE, Kennedy MA, Leslie JL, Chin DL, Wang S, Nguyen HH, Huang B, Tang YW, Lee LW, Kim K, Taylor S, Romano PS, Panacek EA, Goodell PB, Solnick JV, Cohen SH. Overdiagnosis of Clostridium difficile infection in the molecular test era. JAMA Intern Med 2015; 175(11): 1792–1801.
The authors
Guilherme Grossi Lopes Cançado*1,2 MD, Rodrigo Otávio Silveira Silva3 PhD, and Eduardo Garcia Vilela1 MD, PhD
1Instituto Alfa de Gastroenterologia,
Federal University of Minas Gerais, Brazil.
2Department of Gastroenterology,
Hospital da Polícia Militar de Minas Gerais, Brazil.
3Veterinary School, Federal University of Minas Gerais, Brazil.
*Corresponding author
E-mail: guilhermegrossi@terra.com.br