Diagnosis of the intestinal parasite Strongyloides stercoralis by detection of cell-free parasite DNA fragments in urine
Diagnosis of infection with the parasitic roundworm Strongyloides stercoralis is currently done by stool sample culture to detect active larvae. However, the sensitivity of this method can be as low as 28%. This article describes how cell-free parasite DNA can be detected in urine when the results of stool-sample testing are negative.
by Dr Clive Shiff and Dr Alejandro Krolewiecki
Introduction
Of the neglected tropical diseases, Strongyloides stercoralis infection has emerged as a global problem because it is difficult to diagnose and is often silent but long-lived [1]. Currently the definitive test is performed by examining or making a culture of fresh stool to detect active larvae. Serological analysis for specific antibodies is also used but this is far from definitive [2]. This intestinal parasite has an unusual life cycle which is still somewhat enigmatic and can have dire implications if the patient becomes immunosuppressed [3]. This can happen as a patient ages but also if, for some reason, is placed on immunosuppression therapy. To put this in perspective it is important to appreciate the complexity of this infection and the importance of a simple effective diagnostic test.
The infection is caused by parthenogenetic females that live in the upper reaches of the intestinal tract. Unlike hookworms, which also have adults in the gut, Str. stercoralis does not lay eggs which exit in the stool, embryonate and hatch outside the body, Str. stercoralis females incubate the eggs and deposit the eggs in the intestinal mucosa from which the first-stage rhabditiform larvae emerge in large numbers. Some larvae pass in the stool moult, and commence a free-living sexual phase with male and female adults living in the human fecal material. Free-living stages are not parasitic but after several cycles the larvae change from producing the benign, rhabditiform stage to a parasitic filariform stage. These are infective and will penetrate the skin of anyone approaching or contacting the fecal mass. The parasitic stage occurs after the second moult producing the stage called ‘L3’. These larvae secrete proteolytic enzymes and are tissue invasive. However, not all the larvae produced by the parthenogenetic gut parasites are voided in the stool. A proportion of these larvae moult internally and commence the autoinfection stage. They reinvade the host mucosa and reinfect the host and are distributed round the body in the blood and other fluids. In immunocompetent persons these larvae are killed off and their matter is finally excreted through the urine [3]. In people who become immunosuppressed, these larvae continue to survive and accumulate in large numbers and constitute an urgent, life-threatening condition.
Detecting species-specific DNA from urine
Cell-free DNA of parasite origin has been detected in urine of patients with a number of blood-borne and tissue-dwelling parasites. This has been shown with malaria [4], urogenital schistosomes [5], Schistosoma mansoni [6] and others. In all these publications detection of DNA from urine was the most sensitive of serology, parasitological examination of excreta or antigen capture test and the specificity was equal to detection of eggs in excreta [7]. There is also an advantage in using urine specimens. It is simple and can be collected almost on demand. For this work the specimen is filtered through a standard filter paper cone. Approximately 40 ml of urine is filtered, and then the paper is removed from the beaker, opened and allowed to dry in a fly-proof, clean area [8]. When dry each filter is placed in a sealable zip lock plastic bag with a small desiccant capsule. Papers can be stored at 4 °C for months without deterioration of the parasite DNA. In the field when survey work is carried out, urine collection can be carried out simply and in a single day, but filtration and drying of the filters needs to be done within 3 to 4 hours of collection as DNA is degraded by long storage in the urine specimen.
Methods
Ethical clearances
The specimens were collected as part of an ongoing programme to find and cure infections of soil-transmitted helminths by the Ministry of Health and approved by Commité de Ética Colegio Médico de Salta, Salta, Argentina and Johns Hopkins University (IRB number 6199).
Extraction of parasite DNA from filter paper
Filter papers (Whatman No. 3, 12.5 cm diameter) clearly labelled with pencil received in the laboratory are processed as follows. Using a metal punch fifteen 1.00 mm discs are removed from the apex of the quadrant sampled. These are placed in a sterile 1.5 mL Eppendorf tube and 600 µL of nuclease free water added, then incubated at 95 °C for 10 min, and subject to gentle agitation overnight at room temperature. Tubes were then centrifuged at 4000 r.p.m. for 5 min and the supernatant was removed and processed for DNA extraction. We used QIAmpDNA Blood Mini Kit (Qiagen) according to manufacturer’s protocol. The amount of recovered DNA was measured by NanoDrop, ND-1000 spectrophotometer (Thermo Scientific) and stored at −20 °C [9].
Identification of specific Str. stercoralis DNA fragment
Previous work [10] has shown that tandem repeat DNA composed a high proportion of genomic DNA, and these repeats incorporate smaller repeat fragments of DNA. Small fragments of parasite-specific DNA, are found nested within tandem repeats. GenBank AY028262 is such a fragment. Primers for a 125-bp fragment were designed using PrimerQuest Tool (IDT) these are:
Forward (SSC-F) 5´-CTC AGC TCC AGT AAA GCA ACA G-3´
Reverse (SSC-R) 5´-AGC TGA ATC TGG AGA GTG AAG A-3´.
The sequence amplified by these primers was compared with a Blast search against total GenBank data and found only to amplify Str. stercoralis DNA. They were also tested against DNA from three Ancylostoma spp., Sch. mansoni and Sch. haematobium and found only to amplify a product from Str. stercoralis [9].
Amplification and visualization
PCR amplification in 15-µL volume with 2× Taq Mastermix (New England Biolabs), 0.75 µL of 10 µM of each primer, 1–2 µL (20–100 ng/µL) of product DNA made to volume with PCR-grade water (Sigma-Aldrich). The protocol, denaturation at 95 °C to 10 min and 35 cycles at 95 C for 1 min, 63 °C for 1 min 30s, 72 °C for 1 min and a final extension at 72 °C for 10 min. To confirm amplicon size products, were resolved on a 2% agarose gel and stained with Ethidium Bromide (Sigma-Aldrich) [9].
Results
Limits of detection
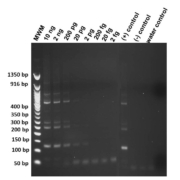
Genomic DNA from Str. stercoralis was diluted and titrated sequentially in concentration from 2 ng/µL to 2 fg/µL to determine the extinction level under standard amplification procedure. Amplifications were performed in duplicate to ensure reproducibility. Products amplified were cleaned with ExoSAP-IT (Afflymetrix Inc.), sequenced and compared with the Str. stercoralis repeat sequence in GenBank (AY028262) to ensure confirmation. In Figure 1 the limit of detection was 20 pg of target DNA.
Diagnostic efficacy
A study was conducted to compare the diagnostic efficacy of parasitological copro-diagnostic methods with DNA detection. For this specimens of stool and urine were collected from 125 individuals living in endemic regions of northern Argentina. The stool specimens were examined fresh using three parasitological tests, concentration- sedimentation, Harada-Mori and Baermann culture methods. Urine samples were filtered as outlined above, dried and sent to the laboratory at Johns Hopkins for DNA extraction and amplification.
The results are given in Table 1 comparing the results of stool versus urine analysis. The prevalence when stool only, 28% is compared with the DNA detection 44.8%, the difference in prevalence is a highly significant 62% difference (P=0.0058). With further analysis comparing the two procedures in the same community, detection of DNA in the urine is more sensitive with significant difference again, 87.5% (95% CL 76.8–94.4) against 56.5% (CL 42.3–69.0%). Specificity in both tests was 100%.
Discussion
There are important reasons for the development of highly sensitive and specific diagnostic tests for the neglected tropical diseases. These relate to modern attempts to limit or eliminate these diseases from much of the endemic areas [11]. However, most parasitic infections have been sustained in their communities for evolutionary time and the parasites have adapted effectively to sustain their populations. This has resulted in very high replicative stages in the life cycle, for instance schistosomes produce large and sustained numbers of cercariae in the snail intermediate hosts from a single miracidium [12]. With strongyloidiasis the multiplication occurs in the host through effective autoinfection; hence, effective control must identify all cases to eliminate the condition. In this work the difference between a prevalence of 28% and 44.8%, means missing almost half of the population at risk. Serological diagnoses are available, but authorities are not satisfied with either the specificity or sensitivity of these tests [13].
The work described there opens an avenue to help ameliorate these problems on two counts. First there is an improvement in sensitivity without loss of specificity, albeit the process requires the use of DNA amplification and detection equipment. This has been mentioned in numerous review articles, but in reality it is an excuse rather than a reason because there are few countries in the world now where there is no access to such equipment. Furthermore the ‘loop mediated amplification procedure’ (LAMP) has been applied to most of these diagnostic methods with success, so amplification is not a real problem. The main difficulty has been in collecting and storing specimens. This has been solved with the use of urine as a vehicle for diagnostic DNA. There are two reasons. First, urine can be obtained on demand; there is no need for a long wait. Second, the specimen is easily collected: the procedure is non-invasive and with simple equipment the sample can be filtered through standard Whatman No. 3 filter paper within minutes of collection. The collection of urine samples for DNA testing has already been done in Nigeria [8] and elsewhere, where colleagues have implemented the work.
Several laboratories are focusing on stool collections as so many soil-transmitted helminths are transmitted by feces. In a hospital environment, collection of a stool sample is a straightforward procedure that can be carried out under clean and safe-handling conditions. DNA detection can be carried out on preserved feces, and using real-time PCR multiplex procedures DNA from various sources (parasitic) can be identified from a single sample and the procedure is currently in use [14]. Although there is an advantage in multiple diagnoses from a single stool, the sensitivity will depend on whether there are actual organisms in the stool examined. In low-density infections, there are times when there is no parasite material in the feces, which will give a false negative response [15]. It has been shown with Sch. mansoni infections, DNA was detected in urine when there were no eggs of the parasite seen in stool [6].
Conclusions
Although this method may not be feasible for all soil-transmitted helminths, detection of parasite-specific DNA in urine seems the best way of achieving optimum sensitivity. The use of urine also has added advantages over stool collection, primarily because it is available more or less on demand, it is simple to handle, does not require fume extraction hoods, it is not dangerous to handle and can be processed in the field, and once collected on dry filter paper it is easily and economically transported.
References
1. Schad G. Morphology and life history of Strongyloides stercoralis. In: Gove DI (Ed) Strongyloidiasis: a major roundworm infection of man. Taylor and Francis 1989.
2. Krolewiecki AJ, Ramanathan R, Fink V, McAuliffe I, Cajal SP, Won K, Juarez M, Di Paolo A, Tapia L, et al. Improved diagnosis of Strongyloides stercoralis using recombinant antigen-based serologies in a community-wide study in northern Argentina. Clin Vaccine Immunol 2010; 17(10): 1624–1630.
3. Schad G, Aikens L, Smith G. Strongyloides stercoralis: is there a canonical migratory route through the host? J Parasitol 1989; 75: 740–749.
4. Mharakurwa S, Simoloka C, Thuma PE, Shiff CJ, Sullivan DJ. PCR detection of Plasmodium falciparum in human urine and saliva samples. Malar J 2006; 5: 103.
5. Ibironke OA, Phillips AE, Garba A, Lamine SM, Shiff C. Diagnosis of Schistosoma haematobium by detection of specific DNA fragments from filtered urine samples. Am J Trop Med Hyg 2011; 84(6): 998–1001.
6. Lodh N, Mwansa JC, Mutengo MM, Shiff CJ. Diagnosis of Schistosoma mansoni without the stool: comparison of three diagnostic tests to detect Schistosoma mansoni infection from filtered urine in Zambia. Am J Trop Med Hyg 2013 July; 89(1): 46–50.
7. Krolewiecki AJ, Lammie P, Jacobson J, Gabrielli AF, Levecke B, Socias E, Arias LM, Sosa N, Abraham D, et al. A public health response against Strongyloides stercoralis: time to look at soil-transmitted helminthiasis in full. PLoS Negl Trop Dis 2013; 7(5): e2165.
8. Ibironke OA, Koukounari A, Asaolu S, Moustaki I, Shiff C. Validation of a new test for Schistosoma haematobium based on detection of the Dra1 DNA repeat fragment in urine: evaluation through latent class analysis. PLoS Negl Trop Dis 2012; 6(1): e1464.
9. Lodh N, Caro N, Sofer S, Scott A, Krolewiecki A, Shiff C. Diagnosis of Strongyloides stercoralis: detection of parasite-derived DNA in urine. Acta Tropica 2016; 163: 9–13.
10. Hamburger J, Turetski T, Kapeller I, Deresiewicz R. Highly repeated short DNA sequences in the genome of Schistosoma mansoni recognized by a species-specific probe. Mol Biochem Parasitol 1991; 44: 73–80.
11. Lo NC, Addiss DG, Hotez PJ, King CH, Stothard R, Evans DS, Colley DG, Lin W, Coulibaly JT, et al. A call to strengthen the global strategy against schistosomiasis and soil-transmitted helminthiasis: the time is now. Lancet Infect Dis 2016.
12. Shiff C. The importance of definitive diagnosis in chronic schistosomiasis, with reference to Schistosoma haematobium. J Parasitol Res 2012; 2012: 761269.
13. Bisoffi Z, Buonfrate D, Sequi M, Mejia R, Cimino RO, Krolewiecki AJ, Albonico M, Gobbo M, Bonafini S, et al. Diagnostic accuracy of five serologic tests for Strongyloides stercoralis infection. PLoS Negl Trop Dis 2014; 8(1): e2640.
14. Basuni M, Muhi J, Othman N, Verweij JJ, Ahmad M, Miswan N, Rahumatullah A, Aziz FA, Zainudin NS, Noordin R. A pentaplex real-time polymerase chain reaction assay for detection of four species of soil-transmitted helminths. Am J Trop Med Hyg 2011; 84(2): 338–343.
15. Lodh N, Naples JM, Bosompem KM, Quartey J, Shiff CJ. Detection of parasite-specific DNA in urine sediment obtained by filtration differentiates between single and mixed infections of Schistosoma mansoni and S. haematobium from endemic areas in Ghana. PLoS One 2014; 9: e91144.
The authors
Clive Shiff*1 PhD and Alejandro Krolewiecki2 MD, PhD
1Department of Molecular
Microbiology and Immunology,
Johns Hopkins Bloomberg
School of Public Health,
Baltimore, MD 21205, USA
2Instituto de Investigaciones en
Enfermedades Tropicales,
San Ramón de la Nueva Orán 4530,
Salta, Argentina
*Corresponding author
E-mail: cshiff1@jhu.edu