Next-generation sequencing in clinical virology diagnostics
Introduction
Methodologies to detect pathogenic viruses in clinical specimens have transitioned from classic cell culture and antibody–antigen techniques to more sensitive molecular methods such as polymerase chain reaction (PCR). The targeted nature of these methodologies inhibits their ability to accommodate the true diversity of human pathogens in a clinical specimen, especially viruses [1]. Next-generation sequencing (NGS) technologies are quickly demonstrating their ability to provide broad detection of infectious agents in a target-independent manner [2–7]. NGS has many advantages beyond the improved detection of all suspected, unsuspected, or even novel pathogens in a clinical specimen [8]. Familiarization with pathogen genomic sequences within clinical specimens enhances our understanding of infectious disease through further discovery of pathogen variability and genotyping [9–11], drug resistance or response to therapy [12], vaccine development and efficacy monitoring [13], and further characterization of the metagenome [14]. The use of NGS for routine use in clinical diagnostics is emerging with its own set of limitations and challenges [13, 15]. Focusing on viruses of public health importance, we compared the performance of NGS alongside other more common viral detection methodologies.
Conventional methods versus NGS
We investigated applications of NGS in a clinical laboratory to detect pathogenic viruses in common specimen types and compared NGS data to that which could be obtained by more conventional methods for detecting and characterizing the following viruses of public health importance: adenovirus, herpesvirus, hepatitis C virus, and influenza [16]. We compared results obtained by NGS to viral culture, immunofluorescence staining, serum neutralization, and PCR in terms of turnaround time as well as the clinical relevance of the information obtained.
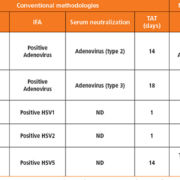
Table 1 describes the turnaround time of conventional methods to NGS for detecting adenoviruses and herpesviruses, both DNA viruses. The amount of time it takes to grow a virus in culture is variable, ranging from 1 day for herpes simplex viruses to 18 days for adenoviruses. All NGS data could be obtained in 4 days, which includes nucleic acid extraction, sequencing library preparation, sequencing and data analysis. Although most laboratories are not currently equipped with in-house bioinformaticians, much of the analysis can be done simply using common sequencing analysing software and the quickly growing number of applications online. For data analysis, we used PathSeq™Virome which enabled us to feed large read files into the application which would generate a report describing the viruses present, including a ‘detection score’ to distinguish strong and weak presence. NGS data provided much more information regarding the exact isolate which may aid health professionals in tracking and relating individual cases with others. Group C adenoviruses are treatable with cidofovir and NGS data was able to identify the amino acid motif that most affects antiviral resistance.
Hepatitis C virus (HCV) is a growing concern for public health and tends to be difficult to design targeted methodologies around owing to the high variability of viral genomes known, even within the same patient. NGS is a powerful tool for characterizing HCV infections and, in our experience, more informational than targeted genotyping assays (Table 2). As we were able to sequence nearly the entire HCV genome (coverage ranged from 92.4–95.6%), data could be generated describing the mutations at key locations across the genomes that are known to cause drug resistance. Antiviral resistance is also critical when characterizing current circulating influenza virus strains and NGS was able to identify viruses that would be considered susceptible to neuraminidase inhibitors (Table 3). In two cases, the viral load of the specimen was too low to achieve good genome coverage across the neuraminidase gene, but this issue could be resolved by screening specimens for high titre (i.e. qPCR) or using enrichment techniques such as ultracentrifugation or filtration of other background nucleic acid.
In most cases, with the exception of one specimen where no cytomegalovirus was definitively identified (HSV5, Table 1), information retrieved by NGS met or exceeded that of conventional methodologies. NGS proves to be a laboratory tool capable of not only detecting pathogenic viruses in clinical specimens, but also predicting the effects of drug treatment as well.
Summary
Through increased use of NGS technologies, reference databases of whole genome sequences can grow and enhance our ability to more definitively identify sequencing reads. Although this review describes conventional methods versus NGS for detecting specific viruses, there was also evidence of the presence of co-infecting viruses such as hepatitis G and Torque Teno virus that weren’t originally targeted. The standard 4-day turnaround time needed to complete NGS could be improved with extraction and library preparation automation, as well as advances in sequencing technology (each run ~40 hours). Based on our laboratory’s experience and the growing body of literature, NGS will change our approach as clinical laboratorians and improve our ability to detect and more fully characterize agents of infectious disease in clinical specimens in a non-targeted manner.
References
1. Köser CU, Ellington MJ, Cartwright EJ, Gillespie SH, Brown NM, Farrington M, Holden MT, Dougan G, Bentley SD, Parkhill J, Peacock SJ. Routine use of microbial whole genome sequencing in diagnostic and public health microbiology. PLoS Pathogens 2012; 8(8): e1002824.
2. Bzhalava D, Johansson H, Ekström J, Faust H, Möller B, Eklund C, Nordin P, Stenquist B, Paoli J, Persson B, Forslund O, Dillner J. Unbiased approach for virus detection in skin lesions. PLoS One 2013; 8(6): e65953.
3. Cheval J, Sauvage V, Frangeul L, Dacheux L, Guigon G, Dumey N, Pariente K, Rousseaux C, Dorange F, Berthet N, Brisse S, Moszer I, Bourhy H, Manuguerra CJ, Lecuit M, Burguiere A, Caro V, Eloit M. Evaluation of high-throughput sequencing for identifying known and unknown viruses in biological samples. J Clin Microbiol 2011; 49(9): 3268–3275.
4. Chan BK, Wilson T, Fischer KF, Kriesel JD. Deep sequencing to identify the causes of viral encephalitis. PLoS One 2014; 9(4): e93993.
5. Kriesel JD, Hobbs MR, Jones BB, Milash B, Nagra RM, Fischer KF. Deep sequencing for the detection of virus-like sequences in the brains of patients with multiple sclerosis: detection of GBV-C in human brain. PLoS One 2012; 7(3): e31886.
6. Moore RA, Warren RL, Freeman JD, Gustavsen JA, Chénard C, Friedman JM, Suttle CA, Zhao Y, Holt RA. The sensitivity of massively parallel sequencing for detecting candidate infectious agents associated with human tissue. PLoS One 2011; 6(5): e19838.
7. Yozwiak NL, Skewes-Cox P, Stenglein MD, Balmaseda A, Harris E, DeRisi JL. Virus identification in unknown tropical febrile illness cases using deep sequencing. PLoS Negl Trop Dis 2012; 6(2): e1485.
8. Radford AD, Chapman D, Dixon L, Chantrey J, Darby AC, Hall N. Application of next-generation sequencing technologies in virology. J Gen Virol 2012; 93(9): 1853–1868.
9. Arroyo LS, Smelov V, Bzhalava D, Eklund C, Hultin E, Dillner J. Next generation sequencing for human papillomavirus genotyping. J Clin Virol 2013: 58(2): 437–442.
10. Flaherty P, Natsoulis G, Muralidharan O, Winters M, Buenrostro J, Bell J, Brown S, Holodniy M, Zhang N, Ji HP. Ultrasensitive detection of rare mutations using next-generation targeted resequencing. Nucleic Acids Res 2012; 40(1): e2.
11. Meiring TL, Salimo AT, Coetzee B, Maree HJ, Moodley J, Hitzeroth II, Freeborough M-J, Rybicki EP, Williamson AL. Next-generation sequencing of cervical DNA detects human papillomavirus types not detected by commercial kits. Virol J 2012; 9: 164.
12. Sijmons S, Van Ranst M, Maes P. Genomic and functional characteristics of human cytomegalovirus revealed by next-generation sequencing. Viruses 2014; 6(3): 1049–1072.
13. Watson SJ, Welkers MRA, DePledge DP, Coulter E, Breuer JM, de Jong MD, Kellam P. Viral population analysis and minority-variant detection using short read next-generation sequencing. Philos Trans R Soc Lond B Biol Sci 2013; 368(1614): 20120205.
14. Han Y, Zhang Y, Mei Y, Wang Y, Liu T, Guan Y, Tan D, Liang Y, Yang L, Yi X. Analysis of hepatitis B virus genotyping and drug resistance gene mutations based on massively parallel sequencing. J Virol Methods 2013; 193(2): 341–347.
15. Messiaen P, Verhofstede C, Vandenbroucke I, Dinakis S, Van Eygen V, Thys K, Winters B, Aerssens J, Vogelaers D, Stuyver LJ, Vandekerckhove L. Ultra-deep sequencing of HIV-1 reverse transcriptase before start of an NNRTI-based regimen in treatment-naive patients. Virology 2012; 426(1): 7–11.
16. Parker J, Chen J. Application of next generation sequencing for the detection of human viral pathogens in clinical specimens. J Clin Virol 2017; 86: 20–26.
The authors
Jayme Parker1,2 PhD and Jack Chen*1,2 PhD
1Department of Biology and Wildlife, Institute of Arctic Biology, University of Alaska Fairbanks, Fairbanks, AK 99775, USA
2Alaska State Public Health Virology Laboratory, Fairbanks, AK 99775, USA
*Corresponding author
E-mail: j.chen@alaska.edu