Biomarkers in age-related macular degeneration
Age-related macular degeneration is a late-onset disease of the eye macula that can result in blindness and in a significant deterioration of quality of life. Genetics and oxidative stress from light exposure and smoking are major risk factors. In this brief report, we discuss genetic and plasma epigenetic biomarkers that are examined for their association with the disease.
by Prof. Christos Kroupis, Prof. George Kitsos, Prof. Marilita M. Moschos and Prof. Michael B. Petersen
Introduction
Age-related macular degeneration (AMD) is a slow and progressive disease of the macula, i.e. the central part of the retina, and the leading cause of irreversible visual loss in the Western world. Globally, AMD accounts for 8.7% of all blindness and is predicted to affect 196 million people by 2020; it is more prevalent in populations of European descent than those of Asian and African descent [1]. With the loss of central vision frequently involving both eyes, AMD is a debilitating condition affecting daily tasks such as reading and driving, and ultimately having severe consequences on independence and quality of life. AMD is a late-onset disease with a complex etiology. Major risk factors contributing to susceptibility include age, family history (genetics), light exposure and smoking [2–4].
AMD can be considered a multifactorial dysfunction of the retinal photoreceptor cells and their support system, which includes the retinal pigment epithelium (RPE), Bruch’s membrane (BrM), and the choroidal vasculature. The fundamental cause of vision loss in AMD is the progressive damage to photoreceptors, which can be triggered by RPE dysfunction and atrophy, impaired transport of oxygen, nutrients and metabolites between vessels and outer retinal cells and leakage from choroidal capillaries that invade the retina through the RPE [5].
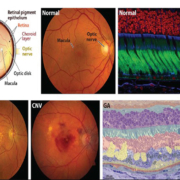
Light entering the eye is focused on the retina, where delicately specialized rod and cone photoreceptors allow its transduction into chemical signals to visual centers in the brain. Photoreceptors are metabolically active neurons with oxygen requirements that are among the highest in the human body. In humans, rods and cones exhibit a distinct topography; the macula (6-mm diameter) contains a cone-dominated fovea (0.8-mm diameter) that is associated with high-acuity vision [5] (Fig. 1a). Just posterior to the photoreceptors, the RPE consists of polarized epithelial cells located at the base of the retina as a single layer of hexagonal cells that are densely packed with pigment granules (melanosomes). The RPE is firmly attached to the underlying basement membrane (BrM). The RPE provides the nutrients needed to maintain visual function by light-sensitive outer segments of the photoreceptors. RPE melanosomes absorb excess incoming light, which protects the retina from light damage. Other critical roles for the RPE involve phagocytosing shed outer retinal segments and scavenging photoreceptor debris, thus, serving as part of the waste-disposal system for the retina. The RPE is known to produce and to secrete a variety of growth factors to help build and sustain the choroid and photoreceptors [6]. The choroid is an extensive vascular meshwork of capillaries lining the posterior part of the eye that supplies nutrients utilized by the retina and acts as a conduit for the by-products of photoreceptor and RPE metabolism [5]. The inner aspect of the choroid, next to the RPE, is the BrM, a laminar extracellular matrix composed mainly of collagen and elastin. Accumulating evidence suggests that the molecular, structural and functional properties of the BrM are dependent on age, genetics, environmental factors, retinal location and disease state. As a result, some properties of the BrM are unique to each human individual at a given age and, therefore, affect uniquely the progression of AMD [6].
AMD pathology
There are two AMD forms: dry (in 90% of patients) and wet (in 10%). In the dry form of AMD, apoptosis of the RPE, neuroretina and choriocapillaris progresses slowly and causes permanent central vision loss. Initially, the BrM exhibits increased deposition of cholesterol and calcium with age. Drusen genesis is a sign of AMD progression (Fig. 1b). Drusen are amorphic extracellular deposits of lipids, proteins, inflammatory molecules in the space between RPE and BrM. The alternative complement path is activated by lipofuscin constituents (which are mostly by-products of the retinal vision cycle) as a response to the inflammatory process connected with drusen genesis. Unfortunately, as we age, mitochondrial function decreases (and mtDNA mutations accumulate) and, therefore, oxidative damage increases. In parallel, antioxidant capacity decreases and the efficiency of repair systems and cytoprotective ubiquitin proteolytic system become impaired [4]. Environmental factors associated with increased production of reactive oxygen species (ROS), such as increased light exposure and cigarette smoking, are additive and have been linked with AMD risk. Collectively, these factors create an environment in which proteins, DNA and lipids become oxidatively damaged. The combination of inadequately neutralized oxidized proteins in the drusen and inflammation associated with OSEs (oxidative specific epitopes) induce focal loss of RPE cells, degeneration of the overlying photoreceptors and vision loss as described in Figure 2 [4].
In the advanced dry form of AMD, geographic atrophy (GA) develops from large, confluent drusen proceeds to hyperpigmentation and then, to cell apoptosis. At present, there is no effective treatment of the dry form. In the wet form, the cause of potential central vision loss is choroidal neovascularization (CNV). An inflammatory reaction initiates pathological angiogenesis that penetrates through defects in the BrM and the RPE layers to the subretinal space, where exudation and bleeding destroy photoreceptors. Commonly used anti-VEGF factors given in repeated intravitreal injections inhibit neovascularization and can stabilize vision acuity in most wet AMD patients.
Genetic biomarkers in AMD
Identification of associated genetic variants can help uncover disease mechanisms and provide entry points for therapy. Linkage of AMD families to 1q32 and the complement factor H (CFH) gene by many groups in 2005, led to the identification of the first common genome-wide significant risk variant, Y402H (rs1061170, g.43097C>T) with variable frequencies across various populations. This SNP (single nucleotide polymorphism) results in an impaired alternative complement pathway inactivation. This discovery propagated numerous genetic and genomic studies that have contributed to our understanding of the pathological mechanisms contributing to AMD. Notably, the subsequent association of common and rare alleles at or near several additional complement genes (CFH, C2/CFB, C3, CFI and C9) has led to the ‘inflammation hypotheses’, with cumulative evidence from genetics and histopathological studies [3]. Another major non-complement pathway AMD-associated locus lies on chromosome 10q26 (LOC387715) and many studies have demonstrated a strong association between AMD and the ARMS2 gene that encodes for a small 107-amino acid protein. ARMS2 A69S SNP (rs10490924, g.5270G>T) is a mutation associated with subsequent mitochondrial dysfunction, ROS generation and accumulation of somatic mitochondrial DNA mutations. These initial promising findings prompted world-wide efforts and culminated in the AMD Gene consortium 2013 study where 19 common variants were associated with the disease in a large number of patients with the use of SNP microarrays; still the two aforementioned SNPs possessed the highest odds ratios (OR) for AMD development (between 2.4 and 2.7) with some differences in their effect according to their different allele frequencies in various populations. It was estimated that these 19 variants can explain ~45% of the genetic heterogeneity in AMD patients above 85 years old; the two main AMD associations with CFH and ARMS2 genes account for a significant 25% of the total cases [5]. Therefore, we and other groups have developed fast, high-throughput robust and accurate genotyping assays for their accurate detection (Fig. 3) [7–9]. Early identification of individuals at risk provides an opportunity to prevent or attenuate the AMD disease. Homozygosity for both CFH and ARMS2 risk alleles increases the progression to advanced AMD stages (GA or CNV) to 48% compared to 5% for those carrying wild-type alleles in both genes [10]. Models incorporating these alleles and/or an expanded variant panel along with smoking and body mass index have been the basis for various commercial tests estimating AMD risk, such as RetnaGene (Nicox), Macula/Vita Risk (ArcticDx), Asper Ophthalmics, etc. Potential nutrigenetic antioxidant interventions have been proposed based on CFH and ARMS2 genotypes [11, 12]. In dry AMD where no therapy exists, anti-complement antibodies are in clinical trials right now (eculizumab, lampalizumab) and genetic tests providing information for complement polymorphisms could select appropriate patients that could benefit from such therapy.
The largest and latest 2016 AMD Gene Consortium study identified additional loci by using an Illumina human core exome array for >12 million variants in 16,144 advanced AMD patients versus 17,832 controls; 52 independently associated common and rare variants were distributed across 34 loci [13]. Now that technological advances permit – with the advent of next-generation sequencing platforms – it would be extremely useful to validate AMD-specific gene-panels for these patients.
Plasma epigenetic biomarkers in AMD
A small, non-coding micro(mi)RNA (18–24 nt) binds to specific mRNAs – depending on its sequence – and results in their degradation by cleavage, translational repression and/or polyA-deadenylation. One miRNA can target many mRNAs but also one mRNA can be targeted by many miRNAs. Emerging evidence arising from tissue studies suggest that beside environmental and genetic factors, epigenetic mechanisms (such as miRNA regulation of gene expression) are relevant to AMD and are providing an exciting new avenue for research and therapy. Sera and plasma (which are easily collected non-invasively) contain cell free DNA, RNA and circulating nucleic acids that can serve as potential biomarkers. The miRNAs identified in human plasma are known to be relatively stable, as they have been found to be resistant to RNase degradation. A recent study has identified a plasma miRNA expression profile specific for AMD patients [14]. Plasma miRNA expression was first screened for multiple miRNAs and then, those showing differences between patients and healthy controls were further explored with individual, specific RT-qPCR assays in a larger number of samples. In another study exploring wet and dry AMD differences in plasma, the miRNA expression analysis revealed increased expression of miR661 and miR3121 in dry AMD patients and miR4258, miR889 and let7 in wet AMD patients compared to controls [15].
References
1. Wong WL, Su X, Li X, Cheung CM, Klein R, Cheng CY, Wong TY. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob Health 2014; 2: e106–e116.
2. Kokotas H, Grigoriadou M, Petersen MB. Age-related macular degeneration: genetic and clinical findings. Clin Chem Lab Med 2011; 49: 601–616.
3. Tan PL, Bowes RC, Katsanis N. AMD and the alternative complement pathway: genetics and functional implications. Hum Genomics 2016; 10: 23.
4. Chiras D, Kitsos G, Petersen MB, Skalidakis I, Kroupis C. Oxidative stress in dry age-related macular degeneration and exfoliation syndrome. Crit Rev Clin Lab Sci 2015; 52: 12–27.
5. Fritsche LG, Fariss RN, Stambolian D, Abecasis GR, Curcio CA, Swaroop A. Age-related macular degeneration: genetics and biology coming together. Annu Rev Genomics Hum Genet 2014; 15: 151–171.
6. Bhutto I, Lutty G. Understanding age-related macular degeneration (AMD): relationships between the photoreceptor/retinal pigment epithelium/Bruch’s membrane/choriocapillaris complex. Mol Aspects Med 2012; 33: 295–317.
7. Velissari A, Skalidakis I, Oliveira SC, Koutsandrea C, Kitsos G, Petersen MB, Kroupis C. Novel association of FCGR2A polymorphism with age-related macular degeneration (AMD) and development of a novel CFH real-time genotyping method. Clin Chem Lab Med 2015; 53: 1521–15219.
8. Sarli A, Skalidakis I, Velissari A, Koutsandrea C, Stefaniotou M, Petersen MB, Kroupis C, Kitsos G, Moschos MM. Investigation of associations of ARMS2, CD14, and TLR4 gene polymorphisms with wet age-related macular degeneration in a Greek population. Clin Ophthalmol 2017; 11: 1347–1358.
9. Xu Y, Guan N, Xu J, Yang X, Ma K, Zhou H, Zhang F, Snellingen T, Jiao Y, et al. Association of CFH, LOC387715, and HTRA1 polymorphisms with exudative age-related macular degeneration in a northern Chinese population. Mol Vis 2008; 14: 1373–1381.
10. Seddon JM, Francis PJ, George S, Schultz DW, Rosner B, Klein ML. Association of CFH Y402H and LOC387715 A69S with progression of age-related macular degeneration. JAMA 2007; 297: 1793–1800.
11. Awh CC, Hawken S, Zanke BW. Treatment response to antioxidants and zinc based on CFH and ARMS2 genetic risk allele number in the Age-Related Eye Disease Study. Ophthalmology 2015; 122: 162–169.
12. Vavvas DG, Small KW, Awh CC, Zanke BW, Tibshirani RJ, Kustra R. CFH and ARMS2 genetic risk determines progression to neovascular age-related macular degeneration after antioxidant and zinc supplementation. Proc Natl Acad Sci U S A 2018; 115: E696–E704.
13. Fritsche LG, Igl W, Bailey JN, Grassmann F, Sengupta S, Bragg-Gresham JL, Burdon KP, Hebbring SJ, Wen C, et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet 2016; 48: 134–143.
14. Ertekin S, Yıldırım O, Dinç E, Ayaz L, Fidancı SB, Tamer L. Evaluation of circulating miRNAs in wet age-related macular degeneration. Mol Vis 2014; 20: 1057–1066.
15. Szemraj M, Bielecka-Kowalska A, Oszajca K, Krajewska M, Goś R, Jurowski P, Kowalski M, Szemraj J. Serum microRNAs as potential biomarkers of AMD. Med Sci Monit 2015; 21: 2734–2742.
The authors
Christos Kroupis*1 MSc, PhD; George Kitsos2 MD, PhD; Marilita M. Moschos3 MD, PhD; Michael B. Petersen4 MD, PhD
1Department of Clinical Biochemistry and Molecular Diagnostics, Attikon University General Hospital, Medical School, National and Kapodistrian University of Athens, Athens, Greece
2Department of Ophthalmology, University General Hospital of Ioannina, Ioannina, Greece
31st Department of Ophthalmology, “G. Gennimatas” General Hospital, Medical School, National and Kapodistrian University of Athens, Athens, Greece
4Department of Clinical Genetics, Aalborg University Hospital, Aalborg, Denmark
*Corresponding author
E-mail: ckroupis@med.uoa.gr