BRCA and beyond: the genes that influence breast cancer risk
Over the last twenty-five years, breast cancer genetics has moved from linkage in high-risk families to association in population-based studies. Accordingly, the genetic variants that have been identified range from rare high-penetrance mutations to common low-penetrance markers. We summarize current knowledge and consider whether understanding how these that variants influence risk could help to refine risk prediction and develop targeted therapies.
by Dr Olivia Fletcher and Dr Syed Haider
Rare high-penetrance mutations
The earliest evidence for genetic susceptibility to cancer came from epidemiological studies in the 1940s and 1950s showing increased cancer risk in the relatives of cancer patients. It was not until the 1990s that linkage analysis, i.e. the genotyping of genetic markers in large family pedigrees, led to the identification of the first breast cancer susceptibility gene, BRCA1, at 17q21 [1]. Identification of the second breast cancer susceptibility gene, BRCA2, at 13q12-13 followed relatively quickly [2]. Mutations in BRCA1 and BRCA2 are present at a frequency of approximately 1 in 800 for BRCA1 and 1 in 500 for BRCA2, they confer high relative risks of breast cancer in carriers (more than tenfold) and are associated with early onset disease [3, 4].
Moderate-risk variants
The next milestone in breast cancer genetics came in 2002 with the discovery of frameshift alteration in the checkpoint kinase 2 gene, CHEK2*1100delC. This variant was discovered using a combination of linkage and mutation screening in a large multiple-case breast cancer family from the Netherlands, followed by analysis of the CHEK2*110delC variant in high-risk breast cancer families, ‘unselected’ breast cancer cases and controls [5]. The CHEK2*1100delC variant occurs on a single haplotype indicating that all CHEK2*1100delC-carrying chromosomes arise from a single founder; this variant is confined to Northern European populations with a prevalence in controls that varies significantly between Northern European populations. Compared to truncating mutations in BRCA1 and BRCA2, the relative risk associated with CHEK2*1100delC is moderate – approximately twofold.
Subsequent to the discovery of CHEK2*1100delC, additional moderate-risk variants were identified in candidate genes including ataxia telangiectasia mutated (ATM), partner and localiser of BRCA2 (PALB2) and BRCA1 interacting protein C-terminal helicase 1 (BRIP1). These variants were discovered by sequencing of exons and exon/intron boundaries of DNA damage repair genes in breast cancer cases from high- and moderate-risk families. Variants in these genes occur in the population at combined frequencies (per gene) of around 1% and are predominantly protein-truncating mutations.
Common low-penetrance variants
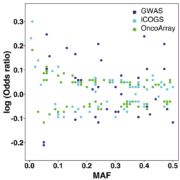
It was not until 2007 that the first genome-wide association study (GWAS) of breast cancer successfully identified five common low-penetrance variants; minor allele frequencies of these variants ranged from 25 to 40% and they were associated with relative risks of 1.07 to 1.26 [6]. Detecting relative risks of this magnitude required three stages of genotyping and a total of 26 258 cases and 26 894 controls. This study was an order of magnitude larger than any previous study marking the beginning of the era of GWAS as well as large consortia. Since 2007 many more breast cancer GWASs have been published, but the major advances in identifying and cataloguing additional low-penetrance variants have come from large collaborative efforts led by the Breast Cancer Association Consortium (http://bcac.ccge.medschl.cam.ac.uk/); in particular two large analyses – the Collaborative Oncological Gene-environment study (COGS) and OncoArray [7, 8]. To date, more than 150 low-penetrance variants conferring relative risks of approximately 0.81–1.35 have been identified. Not surprisingly, the more common variants with the (relatively) more extreme breast cancer odds ratios were identified first, by the GWASs (shown in deep blue, Fig. 1); less common variants and variants with less extreme odds ratios were identified most recently, by the largest pooled analysis, OncoArray (shown in green, Fig. 1).
Contribution to the excess familial relative risk
Breast cancer, like most common cancers, shows familial aggregation; the risk of breast cancer in the first-degree relative of a breast cancer case is about twice that of the risk in the general population [3]. The proportion of this ‘familial relative risk’ that is explained by one or more variants is the metric used to quantify the relative contributions of the different classes of variants – and to estimate the number of variants that have not yet been identified. Relative proportions of all three types of variants are shown in Figure 2; mutations in BRCA1 and BRCA2 account for approximately the same proportion of the familial relative risk as the sum of the common low-penetrance variants.
Differences between coding variants and non-coding variants
One fundamental difference between the high-penetrance mutations in BRCA1 and BRCA2, the moderate-risk variants in DNA damage repair genes and the low-penetrance variants identified by GWAS is that the vast majority of low-penetrance GWAS variants map to non-coding DNA. Estimating the risk of breast cancer for individual BRCA1 and BRCA2 mutation carriers is not trivial; there is some evidence that breast cancer risks differ according to the position of the mutation within the gene [4] and for BRCA2, there is evidence of effect modification by common low-penetrance variants [9]. For the low-penetrance GWAS variants, however, the problem is rather different; while the relative risks associated with the marker single nucleotide polymorphisms (SNPs) are fairly precisely estimated, the underlying ‘causal’ variants and the genes that these variants influence remain – largely – unknown. Approaches to the functional characterisation of GWAS risk loci include fine-scale mapping of potentially large genomic regions, the analysis of SNP genotypes in relation to expression of nearby genes (eQTL) and the use of chromatin association methods [chromosome conformation capture (3C) and Chromatin Interaction Analysis by Paired-End Tag Sequencing (ChIA-PET)] of regulatory regions to determine the identities of target genes. Regulatory elements have been shown to form physical interactions with the genes that they regulate, often over long distances and frequently ‘skipping over’ proximal genes; chromatin association methods capture these interactions and use them to infer likely target genes. We have recently carried out a high-throughput, high resolution analysis of 63 breast cancer risk loci using Capture Hi-C [10]. We were able to identify 110 putative target genes mapping to 33 risk loci. Although some of these putative target genes were well-known cancer genes others were not; in depth follow-up studies will be required to determine which of these putative target genes truly influence breast cancer risk and the mechanisms by which they do so.
Causal variants and target genes can inform risk prediction and therapy
NICE guidelines for the classification, care and management of breast cancer, based on an individual’s family history of breast and other cancers, are used to classify women into three categories: population risk (<17% lifetime risk), moderate risk (17–30% lifetime risk) and high risk (≥30% lifetime risk; https://www.nice.org.uk/guidance/CG164). The options that are available to a woman – increased surveillance, genetic testing, chemoprevention and prophylactic surgery – depend on which category she falls within. A longer-term aim of GWAS is the development of polygenic risk scores (PRS) that can be incorporated into risk prediction algorithms to refine risk estimates. A recent analysis based on 77 breast cancer-associated SNPs, estimated lifetime risks of breast cancer for women in the lowest and highest quintiles of the PRS as 5.3% (population risk) and 17.2% (moderate risk), respectively [11]. Inclusion of larger numbers of SNPs and incorporating causal variants rather than tag SNPs should improve the discriminatory power of the PRS.
In this era of stratified medicine, identifying the genes that underlie GWAS associations and hence – presumably – contribute to defining disease subgroups, also offers the potential for targeted therapies. For instance, metastatic breast cancer patients with germline BRCA1 or BRCA2 mutations who also lack HER2 expression are eligible for Olaparib [a targeted cancer drug that inhibits poly-ADP ribose polymerase (PARP)] as of January 2018. A recent study demonstrated that Olaparib-treated patients have significantly improved progression-free survival (PFS) compared to patients treated with standard-therapy (median PFS of 7 months vs 4.2 months respectively) [12]. Breast cancer patients with germline BRCA1 or BRCA2 mutations already have a defect in their DNA repair mechanisms; by blocking PARP proteins, Olaparib acts to exacerbate DNA damage and trigger cell death, specifically in cancer cells (synthetic lethality). Although defects in DNA repair can be a consequence of germline BRCA mutations, some breast cancer patients manifest defects in DNA repair in the absence of germline BRCA mutations; these patients are also regarded as BRCA deficient – a characteristic often termed as ‘BRCAness’ [13]. Scientists are actively searching for biomarkers of BRCAness in order to assess the suitability of existing PARP inhibitors for patients exhibiting BRCAness [14]. Additional clinical trials on studying efficacy of PARP inhibitors for treating other breast cancer subgroups are underway.
The associations between GWAS SNPs and disease are very modest, and this is often cited as a disadvantage when it comes to considering the genes that map to these loci as putative drug targets. However, an individual non-coding ‘causal’ SNP will usually explain only a small proportion of variation in expression of the gene(s) that it regulates; chemically targeting these genes could have a much more profound effect on disease incidence or outcome. In support of this prediction, a recent investigation by scientists from GlaxoSmithKline estimated that selecting genetically supported targets (including those identified by GWAS) could double the success rate of drugs in clinical development. Although this estimate may be less applicable to cancer drugs, where the somatic genome is as important – or more important – than the germline genome [15,] it leaves open the possibility of new therapies targeting the genes that underlie GWAS associations.
Acknowledgements
We thank Breast Cancer Now for funding this work as part of Programme Funding to the Breast Cancer Now Toby Robins Research Centre.
References
1. Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 1994; 266(5182): 66–71.
2. Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature 1995; 378(6559): 789–792.
3. Easton DF. How many more breast cancer predisposition genes are there? Breast Cancer Res 1999; 1(1): 14–17.
4. Kuchenbaecker KB, Hopper JL, Barnes DR, Phillips KA, Mooij TM, Roos-Blom MJ, Jervis S7, van Leeuwen FE5, Milne RL, et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA 2017; 317(23): 2402–2416.
5. Meijers-Heijboer H, van den Ouweland A, Klijn J, Wasielewski M, de Snoo A, Oldenburg R, Hollestelle A, Houben M, Crepin E, et al. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet 2002; 31(1): 55–59.
6. Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 2007; 447(7148): 1087–1093.
7. Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, Schmidt MK, Chang-Claude J, Bojesen SE, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet 2013; 45(4): 353–361e2.
8. Michailidou K, Lindstrom S, Dennis J, Beesley J, Hui S, Kar S, Lemaçon A, Soucy P, Glubb D, et al. Association analysis identifies 65 new breast cancer risk loci. Nature 2017; 551(7678): 92–94.
9. Gaudet MM, Kirchhoff T, Green T, Vijai J, Korn JM, Guiducci C, Segrè AV, McGee K, McGuffog L, et al. Common genetic variants and modification of penetrance of BRCA2-associated breast cancer. PLoS Genet 2010; 6(10): e1001183.
10. Baxter JS, Leavy OC, Dryden NH, Maguire S, Johnson N, Fedele V, Simigdala N, Martin LA, Andrews S, et al. Capture Hi-C identifies putative target genes at 33 breast cancer risk loci. Nat Commun 2018; 9(1): 1028.
11. Mavaddat N, Pharoah PD, Michailidou K, Tyrer J, Brook MN, Bolla MK, Wang Q, Dennis J, Dunning AM, et al. Prediction of breast cancer risk based on profiling with common genetic variants. J Natl Cancer Inst 2015; 107(5): pii: djv036.
12. Robson M, Im SA, Senkus E, Xu B, Domchek SM, Masuda N, Delaloge S, Li W, Tung N, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Eng J Med 2017; 377(6): 523–533.
13. Lord CJ, Ashworth A. BRCAness revisited. Nat Rev Cancer 2016; 16(2): 110–120.
14. Davies H, Glodzik D, Morganella S, Yates LR, Staaf J, Zou X, Ramakrishna M, Martin S, Boyault S, et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med 2017; 23(4): 517–525.
15. Nelson MR, Tipney H, Painter JL, Shen J, Nicoletti P, Shen Y, et al. The support of human genetic evidence for approved drug indications. Nat Genet 2015; 47(8): 856–860.
The authors
Olivia Fletcher* PhD, Syed Haider PhD
Breast Cancer Now Toby Robins Research Centre, The Institute of Cancer Research, London SW3 6JB, UK
*Corresponding author
E-mail: Olivia.fletcher@icr.ac.uk