Methodology for finding and interpreting efficient biomarker signals
Modern ‘omics’ and screening technologies make possible the analysis of large numbers of proteins with the aim of finding biomarkers for individually tailored diagnosis and prognosis of disease. However, this goal will only be reached if we are also able to sensibly sort through the huge amounts of data that are generated by these techniques. This article discusses how data analysis techniques that have been developed and refined for over a century in the field of psychology may also be applicable and useful for the identification of novel biomarkers.
by Dr J. Michael Menke and Dr Debosree Roy
Introduction
The profession and practice of medicine are rapidly moving towards more specialization, more focused diagnoses and individualized treatments. The result will be called personalized medicine. Presumably genetic predisposition will remain the primary biological basis, but diagnosis and screening will also evolve from complex system outputs observed as increases or decreases of levels of biomarkers in human secretions and excretions. In this sense, the exploration in the human sciences will undoubtedly expand to new frontiers, interdisciplinary cooperation, new disease reclassifications, and the disappearance of entire scientific professions.
Big data and massive datasets by themselves can never answer our deepest and most troubling questions about mortality and morbidity. After all, data are dumb, and need to be properly coaxed to reveal their secrets [1]. Without theories, our great piles of data remain uninformative. Big data need to be organized for, and subjected to, theory testing or data fitting to best competing theories [2, 3] to avoid spurious significant differences, conceivably the biggest threat to science in history [4, 5].
Old tools for big data
New demands presented by our ubiquitous data require new inferential methods. We may discover that disease is emergent from many factors working together to create a diagnosis in one person that, in fact, actually has many different causes in another person with the same diagnosis. Perhaps there are new diseases to be discovered. There might be better early detection and treatment. Much like the earliest forms of life on earth, pathology is much more complicated than just the rise of plant and animal kingdoms as taught mid-twentieth century in evolution.
Although new methodologies may meet scientific requirements of big data, tools already in existence may obviate the need to invent new ones. In particular, methods developed by and for psychologists over more than 100 years may already be an answer. Established data organization and analysis have already been developed by psychologists to test theories about nature’s most complex systems of life. Inference and prediction from massive amounts of data from multiple sources might yield more from these ‘fine scalpels’ without the need for brute force analyses, such as tests for statistical differences that look significant in many cases because of systematic bias in population data arising from unmeasured heterogeneity. The development of some of the most applicable psychological tools began in the early 20th century for measuring intelligence, skills and abilities. Thus, these tools have been used and refined for over a century. From psychological science emerged elegant approaches to data analysis and reduction to evaluate persons and populations for test validity, reliability, sensitivity, specificity, positive and negative predictive values, and efficiency. Psychological testing and medical screening share a common purpose: measure the existence and extent of largely invisible or hard to measure ‘latent’ attributes by establishing how various indicators that are attached to the latent trait react to the presence or absence of subclinical or unseen disease. Biomarkers are thus analogues of test questions, with each biomarker expressing information that helps establish the presence or absence of disease and its stage of progression. The analogous process recommended in this paper is simply this: How many and what kind of biomarkers are sufficient to screen for disease?
Biomarkers for whole-person healthcare
Although the use of biomarkers seems to buck the popular trend of promoting whole person diagnosis and treatment, biomarkers per se are actually nothing new. Biomarkers as products of human metabolism and waste have played an important role over centuries of disease diagnosis and prognosis, preceding science and often leading to catastrophic or ineffective results (think of ‘humours’ and ‘bloodletting’ as examples). Today, blood and urine chemistries are routinely used for focusing on a common cause (disease) of a number of symptoms. Blood in the stools, excessive thirst, glucose in urine, colour of eye sclera, round out information attributable to a common and familiar cause crucial for identifying and treating a system or body part. Signs of thirst and frequent urination may be necessary, but not sufficient for diagnosis of diabetes mellitus, yet can lead to quick referral or triage. The broad category of the physiological signs (biomarkers) has extended along with technology to the microscopic and molecular.
Today, the general testing for and collection of biomarkers in bodily fluids is a growing medical research frontier. However, too many, biomarkers can be confused with genes and epigenetic expressions of genes. Small distinctions might uncover the discovery of new genes leading to new definitions of disease, more accurate detection, and more personal treatment.
With the flood of data unleashed by research in these areas, a new and fundamental problem arises: How do we make sense of all these data? For now, professions and the public may be putting their faith in ‘big data’ in order to make biomarkers clinically meaningful and informative. We are in good company with those who remind us that data are dumb and can be misused to support bias, and that lots of poor quality data do not compile good science. At its heart, scientific theories need to be tested and scientific knowledge built in supported increments.
Biomarkers as medical tests
As with any medical test, some biomarkers are more accurate, or more related, to disease presence and absence and therefore are better indicators of underlying disease state. Thus, some biomarkers are more accurate than others; or put another way, biomarkers represent ‘mini-medical’ tests and their levels of contribution to diagnoses and prognoses depend upon random factors, along with sensitivity, specificity, and disease prevalence [6]. Some biomarkers may increase in presence with disease but lower with health, or the opposite – lower concentrations with disease. To complicate matters further, there are probably plenty of mixed signals, i.e. biomarker A is more sensitive than biomarker B, but B is more specific than A. Blending the information acquired by multiple biomarkers needs to be organized and read in a sequence to reduce false signals – positives or negatives – or at least minimize errors based on risk of disease and morbidities and mortalities.
Thus, managing and analysing the flood of biological diagnostic data is not the concern here, but rather its interpretation and clinical application. Balancing biomarker information at the clinical level is the function of translational research. Test-and-measurement (T&M) psychologists have worked on the science of organizing and interpreting individual items as revealing underlying latent constructs for over a century. Through the extremely tedious task of measuring human intelligence, skills and abilities, some already developed T&M tools could help improve the science, accuracy and interpretation of biomarkers [6].
Psychometric properties of biomarkers
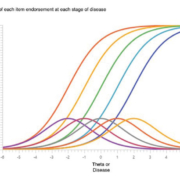
Before embarking on a psychometric approach to biomarker interpretation, some common definitions are required. For instance, what is sensitivity or specificity? A psychometric or medical test shows high sensitivity when the underlying disease or person characteristic is also high. For intelligence, a high-test score implies high intelligence. On a single well-crafted test question, the probability of answering it correctly (formally called probability of endorsing) increases along with higher intelligence; if the question is associated with high intelligence, then the question is a strong or weak indicator of personal intelligence. When many test questions are indicators of intelligence, more correctly endorsed answers of good questions should indicate more intelligence. Indeed, some questions may even be ‘easier’ than others, leading to the need to design questions to fill out the continuum of an underlying intelligence being measured. This procedure is item analysis, a part of item response theory, see Figure 1 for an illustration of how multiple items ‘cover’ a given theta or disease.
Notice how irrelevant is the concept of sensitivity in clinical screening and diagnosis. Sensitivity means that if we already know for sure someone is smart or has a disease, the test and its questions will be correct in describing latent construct (referred to ‘theta’) a certain percentage of the time, based upon the test’s ability to detect and describe the presence or degree of the latent trait. Thus, the proportion of time the question is correct, given that we already know the person’s underlying status, is test or item sensitivity. Sensitivity is a test characteristic given we already know the latent trait – disease status. Symbolically, sensitivity is p(T+|D+), the probability of a positive test score (T+) given we already know the person has the disease (D+). Similarly, specificity is p(T−|D−), the probability of a negative test (T−) or item given that we already know that the patient is confirmed disease-free (D−).
Bayesian induction
Bayes Theorem is useful for many reasons, some controversial. But the conversion of disease prevalence along with biomarker sensitivity and specificity, will axiomatically give the probability of an individual having a disease given a positive test.
In Bayesian terms, the positive predictive value (PPV) is the posterior probability of a patient with a positive test. Two important properties of the PPV are: 1. It is a conversion of population prevalence turned into personal probability of disease based on a person’s positive test; and 2. PPV varies directly with the population prevalence of the disease. One cannot interpret a PPV without starting from its known or estimated population prevalence. PPV decreases with rare disease and increases with common disease, irrespective of tests’ sensitivity or specificity estimates. For further details see Figure 3 in the open access article ‘More accurate oral cancer screening with fewer salivary biomarkers’ by Menke et al. [7].
Sensitivity and specificity are characteristics of the test, not any patient. Such deductive processes are not at all clinically useful. In fact, diagnosing and screening are exactly the inverted probability of that: what is the inferred disease state, D+ or D−, from positive and negative test results? In other words, we want p(D+|T+) instead of p(T+|D+), and p(D−|T−) instead of p(T−|D−). The method for inverting the probabilities from test to patient characteristics is by the application of Bayes’ Theorem. This inverted probability is highly influenced by disease prevalence, however, whereas sensitivity and specificity are not.
Role of prevalence in disease detection
Generally, the higher the disease prevalence in a population, the easier it is to detect. Fortunately, this coincides with good intuitive sense. In fact, when screening for diseases, we need to read the biomarker results diachronically to take advantage of the information added by each biomarker. ‘Diachronically’ refers to reading over time. In the case of biomarker screening, all biomarker antibodies or other detectors of biomarker presence will require the fewest number of biomarkers when read in context of other present biomarkers. Diachronic refers to the order in which biomarkers are read, not the order in which they are administered.
Biomarkers can be strongly or weakly informative. The indicator of strong or weak biomarkers is the diagnostic likelihood ratio, which is shown in the image above.
More explicitly this is called a positive diagnostic likelihood ratio, abbreviated +LR. The higher the +LR, the more information it conveys about the presence or absence of disease. The objective of the inverted probability, p(D+|T+), is called the positive predictive value of a test, PPV.
Diachronic contextual reading
When used in conjunction with other biomarkers, [p(D+|T1, T2, T3, …Tn)], the tests’ accuracy can be increased, but only if the test results are read diachronically. For instance, ‘passing along’ only positive test findings to another biomarker amounts to throwing out true negatives in the sample (and a few false negatives), which increases the ability to detect suspected diseased screened persons from a more prevalent sample pool. After five to ten of these ‘pass-alongs’, depending on original disease prevalence, the PPV can approach 100%, signifying great confidence that a disease is present and further testing and treatment are required. Also, panels of biomarkers – multiple biomarkers used in a single unit for screening – can also have a PPV. In some cases, biomarkers only appear in panels in which case, there is a resultant sensitivity, specificity and PPV for the entire panel.
Biomarkers that are too sensitive might generate too many false positives. This problem can be overcome with a biomarker or biomarkers to ‘clean out’ the false positives. Highly specific biomarkers will throw out false negative ones, a perspective balanced with sensitive biomarkers. Sensitivity and specificity generally vary inversely for each given biomarker. Those high on one attribute tend to be low on the other. Overall, according to our previous experience in meta-analyses, we found specificity was the primary attribute for quickly and accurately screening a population.
The exceptional biomarker can be high on both test attributes. In most cases, the information from mediocre biomarkers can be improved by combining them into biomarker panels with a combined accuracy stronger than any individual biomarker. Once biomarkers are ranked from high to low, wherein they pass along positive test results from highest to lowest dLR, the number of biomarkers required to achieve a PPV near 1.0 is considerably fewer than if biomarkers are ordered from lowest to highest dLR (Fig. 2).
Meta-analysis
As you may have inferred by now, the methodology of identifying the best biomarkers is via meta-analysis. A word of caution for diagnostic meta-analyses. There are software packages for the meta-analysis of medical tests. Meta-DiSc is one such tool [8, 9]. Material on its development may be found here [9]. When last checked, the Meta-DiSc program was being revised to correct some estimate errors and researchers were re-directed to a Cochrane Collaboration page [10]. In short, it is important not to add up all cells as if they represent one large study, because this misrepresents study homogeneity and therefore variance.
We recommend a meta-analysis that uses an index of evidential support [11–13]. In so doing, the weighting of data based on sample size alone may be avoided [7].
Partitioning panels with evidential support estimates
Biomarkers may be either high on sensitivity or specificity. Others may be very high in one attribute, but not the other. Few are high on both. This issue may be overcome by combining a panel made up of the same biomarker(s) of interest, where individual biomarker member weaknesses may be averaged out by including other biomarkers with complementary strengths. A biomarker with high sensitivity and low specificity may be combined with biomarkers of complementary strengths, such as those with low sensitivity and high specificity. The scenario is to combine those biomarkers high in one trait with those high with its complement. This can be tricky as an average accuracy might fall along a diagonal in a receiver operating characteristic (ROC) chart, rendering it a useless test. Indeed, the idea is to maximize the area under the curve on a ROC chart by ‘pulling the curve’ up into the upper left corner to create more area under the curve, representing diagnostic accuracy. For further details see Figure 2 in the open access article ‘More accurate oral cancer screening with fewer salivary biomarkers’ by Menke et al. [7].
The question is whether the combined accuracy is synergistically greater from using two biomarkers or becomes just an arithmetic average of two biomarkers. This conundrum is solved by making sure there are data points in the upper right corner to ‘pull up’ the ROC curve and maximize the area under the curve, which translates roughly to diagnostic accuracy. In fact, sensitivity to cancer or any other disease must be inverted to PPV before the biomarker exhibits utility. Somewhat paradoxically, just using more biomarkers does not increase screening accuracy without being read in the diachronic context of other tests done at the same time. not (again, refer to Fig. 2 in this article).
Should cancer tests detect only binary signals?
From a test and measures perspective, each biomarker is a kind of test question, where the answer to each question is the state of disease in the body. Some questions or biomarkers or biomarker panels are more or less informative because they are more or less sensitive and specific to detecting disease. The answers sought are binary – yes or no. The patient either has a disease or does not. It is up to the properties of the tests to reveal the truth.
As mentioned before, biomarker accuracy varies. No medical test of any kind is 100% accurate. Biomarkers associated with cancer can and do appear at lower levels in healthy individuals. We must understand this principle to decide whether other tests or panels are necessary to improve screening or diagnostic information.
When educational psychologists measure traits and abilities, e.g. IQ, they ask a series of questions. To the degree that the questions are answered ‘correctly’, a person scores higher and has more of the trait or ability to be measured. Creating a survey or questionnaire is a rigorous process. Think of an underlying variable (IQ) as the latent construct. ‘Construct’ is the intended concept we attempt to measure. The construct is not directly measurable, and thus called latent. Each question is a kind of probe that, to various degrees of accuracy, allows indirect observation of the latent construct or disease state. By analogy, biomarkers can be interpreted as test questions indicating the existence of a latent trait or disease.
Pushing the test analogy further, biomarkers might be negatively keyed, i.e. the levels of certain biomarkers are reduced in the presence of disease, or positively keyed with biomarker presence associated with disease. Whereas assessment of traits and abilities measures a continuous scale of latent construct presence, biomarkers answer a simple binary choice: Is the disease present or not?
Biomarker accuracy is estimated by its sensitivity and specificity. Test questions are subject to data reduction techniques (factor analysis), internal consistency within factors, and item response theory to identify redundant questions and design new questions cover gaps in detecting an underlying disease state.
As we are not basic scientists, but rather behavioural and population ones, we cannot address the clinical and laboratory aspects of biomarkers, but in collaboration with colleagues at dental programmes here in Mesa, Arizona and in Malaysia, we came to understand that some biomarkers are more informative than others in screening and diagnosing disease.
Unidimensionality, monotonicity, and local independence properties
Test items should obey conditions of unidimensionality, monotonicity, and local independence. Briefly applied to medical tests, biomarkers should be indicative of the same latent construct (presence of disease), but individual biomarkers should increase (be positive for disease) along with the actual presence of disease [14].
The application of item response theory to academic test scores will reveal that there are gaps in assessment that miss progress or degree of the latent construct. When graphed on person–item maps, the high-ability persons will score higher on the test – i.e. endorse more items, especially the most difficult ones. The item–person map might show two areas of concern: redundant items that may be removed from the test to make the test more efficient, and abilities that cannot be determined owing to items clustering over small ranges of the latent construct. This is exemplified in Figure 1 in Warholak et al. [15].
As for biomarker disease screening, test or panel gaps may miss a subclinical or early stage disease by not matching the stage with biomarkers that would alert us to that stage of disease. In effect, this would be a blind-spot that more research may be required to fill. On the one hand, for a binary screening outcome – yes or no – gaps are not crucial. On the other hand, the discovery of gaps may lead to better science and better early disease detection.
Generalizability theory
Generalizability theory – or G-Theory – is a tool developed by Lee Cronbach and colleagues at Stanford around 1972 [16]. Without getting into excessive detail, it should suffice in this article that G-Theory be mentioned as a methodology for identifying sources of error, bias, or interference in statistical modelling of complex systems. As an example of the reasons for developing G-Theory in the first place, students are taught by professors within classes in courses in schools and states and countries. Each level of this education hierarchy may become a source of variability. If what we want to produce is a similar product in student graduates, as minimal competency in medicine, we may glean interference – variability – introduced by various levels or one specific level. With G-Theory, the primary source of variance may be identified and modified accordingly.
In the biomarker analogy, some biomarkers introduce more confusion than they resolve and can be eliminated or modified to improve reliability and consistent accuracy.
Conclusion
Although biomarker research is being funded and undertaken at unprecedented levels, it is important to remember the credible handling the data in a scientific manner is still the key to understanding and discovery. Big data still needs to answer the question of ‘What does it all mean?’ Yet, we recommend starting with highly refined methodology developed for T&M of human skills, abilities and knowledge. At the very least T&M science might minimize errors, increase medical test efficiencies, and may be used to complement or confirm findings for translational research.
References
1. Pearl J, Mackenzie D. The book of why: the new science of cause and effect. Basic Books 2018.
2. Platt JR. Strong inference: certain systematic methods of scientific thinking may produce much more rapid progress than others. Science 1964; 146(3642): 347–352.
3. Chamberlin TC. The method of multiple working hypotheses. Science 1897, reprint 1965; 148: 754–759.
4. Kline RB. Beyond significance testing: reforming data analysis methods in behavioral research. American Psychological Association 2004.
5. Ziliak ST, McCloskey DN. The cult of statistical significance: how the standard error costs us jobs, justice, and lives. The University of Michigan 2011.
6. Kraemer HC. Evaluating medical tests: objectives and quantitative guidelines. Sage Publications 1992.
7. Menke JM, Ahsan MS, Khoo SP. More accurate oral cancer screening with fewer salivary biomarkers. Biomark Cancer 2017; 9: 1179299X17732007 (https://journals.sagepub.com/doi/full/10.1177/1179299X17732007?url_ver=Z39.88-2003&rfr_id=ori%3Arid%3Acrossref.org&rfr_dat=cr_pub%3Dpubmed#articlePermissionsContainer).
8. Zamora J, Abraira V, Muriel A, Khan K, Coomarasamy A. Meta-DiSc: a software for meta-analysis of test accuracy data. BMC Med Res Methodol 2006; 6: 31.
9. Zamora J, Muriel A, Abraira V. Statistical methods: Meta-DiSc ver 1.4. 2006: 1–8 (ftp://ftp.hrc.es/pub/programas/metadisc/MetaDisc_StatisticalMethods.pdf).
10. Cochrane Methods: screening and diagnostic tests 2018 (https://methods.cochrane.org/sdt/welcome).
11. Goodman SN, Royall R. Evidence and scientific research. Am J Public Health 1988; 78(12): 1568–1574.
12. Menke JM. Do manual therapies help low back pain? A comparative effectiveness meta-analysis. Spine (Phila Pa 1976) 2014; 39(7): E463–472.
13. Royall R. Statistical evidence: a likelihood paradigm. Chapman & Hall/CRC 2000.
14. Beck CJ, Menke JM, Figueredo AJ. Validation of a measure of intimate partner abuse (Relationship Behavior Rating Scale-revised) using item response theory analysis. Journal of Divorce and Remarriage 2013; 54(1): 58–77.
15. Warholak TL, Hines LE, Song MC, Gessay A, Menke JM, Sherrill D, Reel S, Murphy JE, Malone DC. Medical, nursing, and pharmacy students’ ability to recognize potential drug-drug interactions: a comparison of healthcare professional students. J Am Acad Nurse Pract 2011; 23(4): 216–221.
16. Shavelson RJ, Webb NM. Generalizability Theory: a primer. Sage Publications 1991.
The authors
J. Michael Menke* DC, PhD, MA; Debosree Roy PhD
A.T. Still Research Institute, A. T. Still University, Mesa, AZ 85206, USA
*Corresponding author
E-mail: jmenke@atsu.edu